Research
 Computational Robotics
Computational Robotics
Ongoing Projects:
- A Framework for Manipulation Planning and Execution under Uncertainty in Partially-Known Environments
- Scalable Next-Generation Software Infrastructure for High-Dimensional Search
- Collaborative Research: FW-HTF-RM: Robotic Teaching Assistant Systems for Nursing Instructors
- Automated and Robotic Inspection of Flood Control Systems
- Collaborative Research: FW-HTF-R: The Future of Robot-Assisted Nursing: Interactive AI Frameworks for Upskilling Nurses and Customizing Robot Assistance
- A Novel Framework for Informed Manipulation Planning
- Robotic Collaboration through Scalable Reactive Synthesis
- Robot Motion Planning with an Experience Database
- Automating Robot Programming Through Constraint Solving and Motion Planning
- Rethinking Motion Generation for Robots Operating in Human Workspaces
- A Synergistic Multi-Layered Approach for Falsification of Specifications for Hybrid Systems
Other Topics of Interest:
with links to related publications
- Planning from high-level specifications
-
 We have worked extensively on combining high-level reasoning with low-level motion planning. One of the challenges is to create powerful abstractions that help guide the low-level exploration. Another challenge is to provide feedback from the low-level exploration to the high-level planner that can be used to update a high-level plan. We have recently released a software package for integrated task and motion planning called the Task-Motion Kit (TMKit). Lately, we have extended our research to policy synthesis.
We have worked extensively on combining high-level reasoning with low-level motion planning. One of the challenges is to create powerful abstractions that help guide the low-level exploration. Another challenge is to provide feedback from the low-level exploration to the high-level planner that can be used to update a high-level plan. We have recently released a software package for integrated task and motion planning called the Task-Motion Kit (TMKit). Lately, we have extended our research to policy synthesis. - Kinodynamic systems
-
 In kinodynamic planning it is often difficult to find the control inputs that will drive a system from one state to another (even in the absence of obstacles). We have developed several planning algorithms for kinodynamic systems. This includes distributed, asynchronous planning algorithms for multiple car-like systems with second-order dynamics and algorithms for reconfigurable robots with many degrees of freedom (but severe torque limits). The picture above shows a physics simulator used during planning for a reconfigurable robot and the corresponding trajectoruy execution on the actual hardware.
In kinodynamic planning it is often difficult to find the control inputs that will drive a system from one state to another (even in the absence of obstacles). We have developed several planning algorithms for kinodynamic systems. This includes distributed, asynchronous planning algorithms for multiple car-like systems with second-order dynamics and algorithms for reconfigurable robots with many degrees of freedom (but severe torque limits). The picture above shows a physics simulator used during planning for a reconfigurable robot and the corresponding trajectoruy execution on the actual hardware.
- Fundamentals of sampling-based planning
-
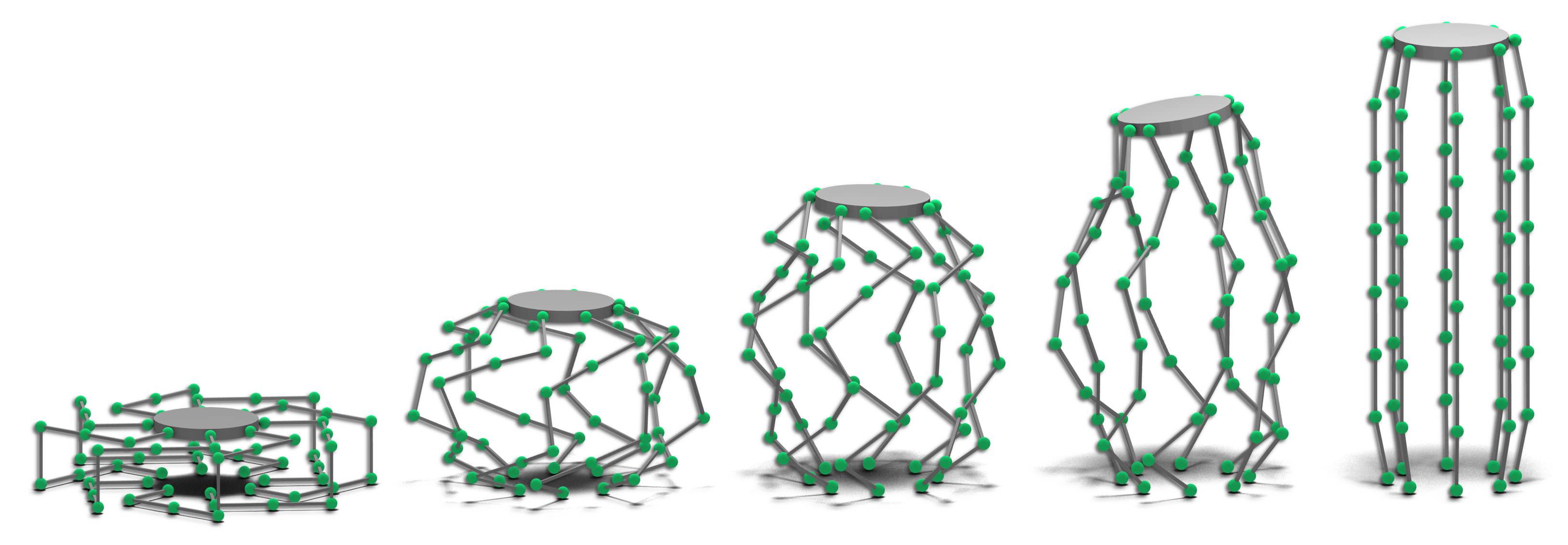 There is a strong emphasis on general algorithms that are not specific to any one particular robot. We have created reference implementations of many of our algrotihms (and many others) that are available as part of the Open Motion Planning Library. The picture above shows an artificial robot with 168 degrees of freedom and many closed chains, which makes it very challenging to plan for. We have recently unified many of the previous approaches to constrained motion planning and generalized it in such a way that almost any sampling-based planner can be used to plan in constrained state space.
There is a strong emphasis on general algorithms that are not specific to any one particular robot. We have created reference implementations of many of our algrotihms (and many others) that are available as part of the Open Motion Planning Library. The picture above shows an artificial robot with 168 degrees of freedom and many closed chains, which makes it very challenging to plan for. We have recently unified many of the previous approaches to constrained motion planning and generalized it in such a way that almost any sampling-based planner can be used to plan in constrained state space.
- Uncertainty
-
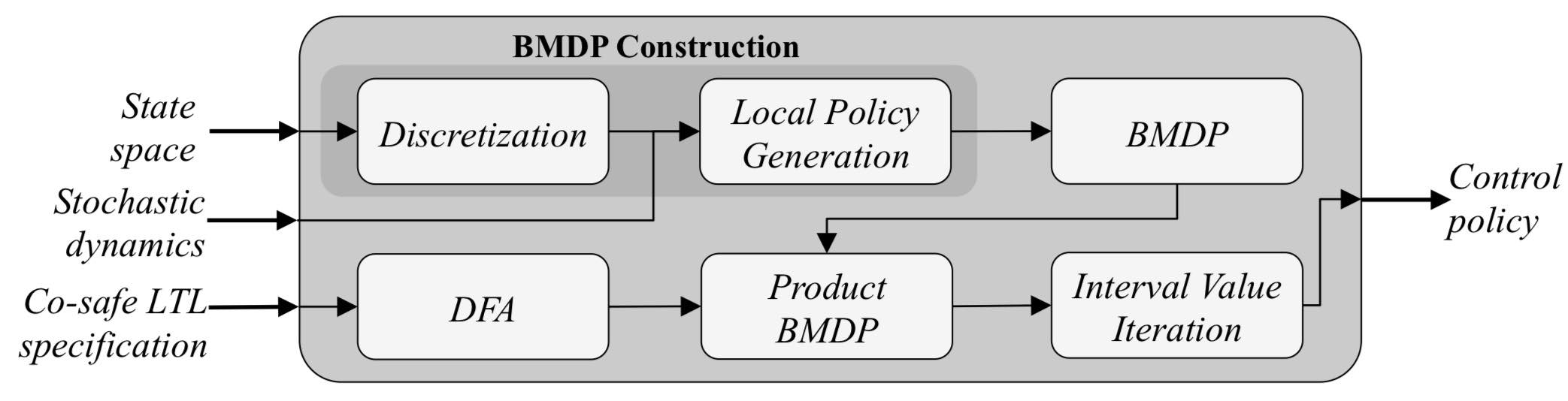 There are different types of uncertainty (action uncertainty, sensing uncertainty, modeling uncertainty). A single plan has in general very little chance of success if it does not account for uncertainty. Solving this problem in its full generality with all types of uncertainty is incredibly difficult, especially if the state, observation and action spaces are all continuous. In many cases, some simplifying assumptions can be made and effective policies for robot motion can still be generated in reasonable time. The picture above illustrates the approach we have taken to synthesize a control policy for a kinodynamic system that realizes a high-level task (specified with a temporal logic) in the presence of action uncertainty.
There are different types of uncertainty (action uncertainty, sensing uncertainty, modeling uncertainty). A single plan has in general very little chance of success if it does not account for uncertainty. Solving this problem in its full generality with all types of uncertainty is incredibly difficult, especially if the state, observation and action spaces are all continuous. In many cases, some simplifying assumptions can be made and effective policies for robot motion can still be generated in reasonable time. The picture above illustrates the approach we have taken to synthesize a control policy for a kinodynamic system that realizes a high-level task (specified with a temporal logic) in the presence of action uncertainty.
- Localization
-
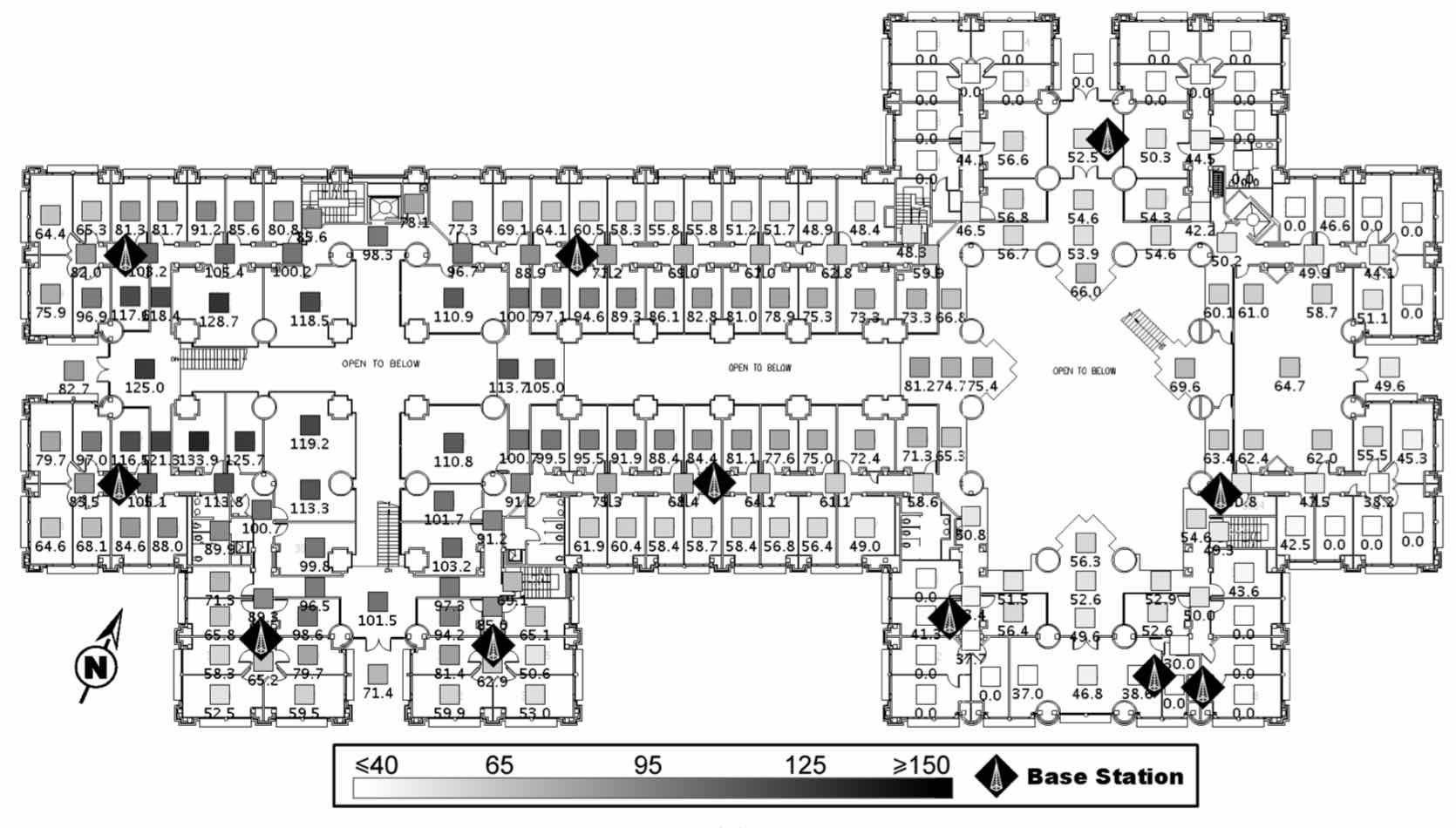 In the early 2000s Simultaneous Localization and Mapping was an extremely active area within robotics. Much of that work relied on expense sensors. We have shown that localization is possible with very simple sensors. For example, in a typical office environment, localization is possible using just WiFi signal strength and the locations of base stations.
In the early 2000s Simultaneous Localization and Mapping was an extremely active area within robotics. Much of that work relied on expense sensors. We have shown that localization is possible with very simple sensors. For example, in a typical office environment, localization is possible using just WiFi signal strength and the locations of base stations.
- Other topics
-
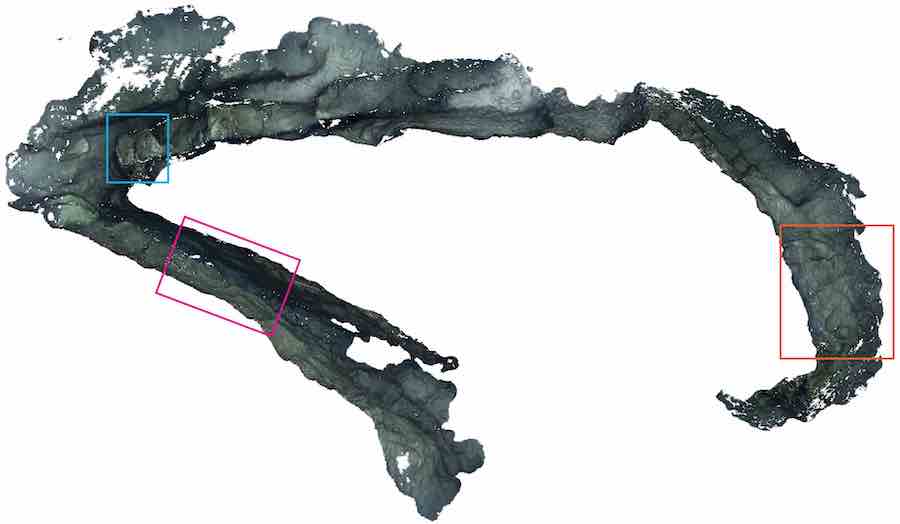 We are interested in a broad range of topics, ranging from mathematical knot untangling to manipulation of deformable objects and active perception. We often have visitors from other groups who also help broaden our horizons. The picture above shows a reconstruction of a canyon using a Autonomous Underwater Vehicle (AUV). This work was performed at the University of Girona. The motion planning capabilities for the AUV were developed in part during visits of Girona students to our group.
We are interested in a broad range of topics, ranging from mathematical knot untangling to manipulation of deformable objects and active perception. We often have visitors from other groups who also help broaden our horizons. The picture above shows a reconstruction of a canyon using a Autonomous Underwater Vehicle (AUV). This work was performed at the University of Girona. The motion planning capabilities for the AUV were developed in part during visits of Girona students to our group.
 Computational Biomedicine
Computational Biomedicine
Ongoing Projects:
- PROTEAN CR: Proteomics Toolkit for Ensemble Analysis in Cancer Research
- Structure-based identification of SARS-derived peptides with potential to induce broad protective immunity
- Structure-based Selection of Tumor-antigens for T-cell Based Immunotherapy
- Structural modeling of peptide-HLA complexes presenting a melanoma-associated antigen for cross-reactivity assessment
- An Integrated Approach to Characterizing Conformational Changes of Large Proteins
- Mining Metabolic and Enzyme Databases for the Composition of Non-canonical Pathways
Other Topics of Interest:
with links to related publications
- Proteins and drugs
-
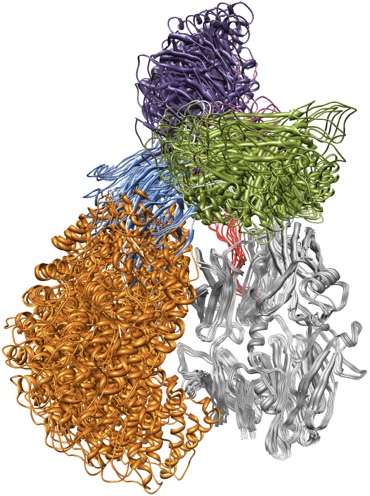 Proteins play a critical role in almost every biological process in the human body. Their function is often mediated by shape changes induced by interactions with other molecules (including drugs). We are working with researchers in the Texas Medical Center to develop methods that allow us to model such conformational changes as well as predict the binding modes of small molecules for a given protein.
Proteins play a critical role in almost every biological process in the human body. Their function is often mediated by shape changes induced by interactions with other molecules (including drugs). We are working with researchers in the Texas Medical Center to develop methods that allow us to model such conformational changes as well as predict the binding modes of small molecules for a given protein.
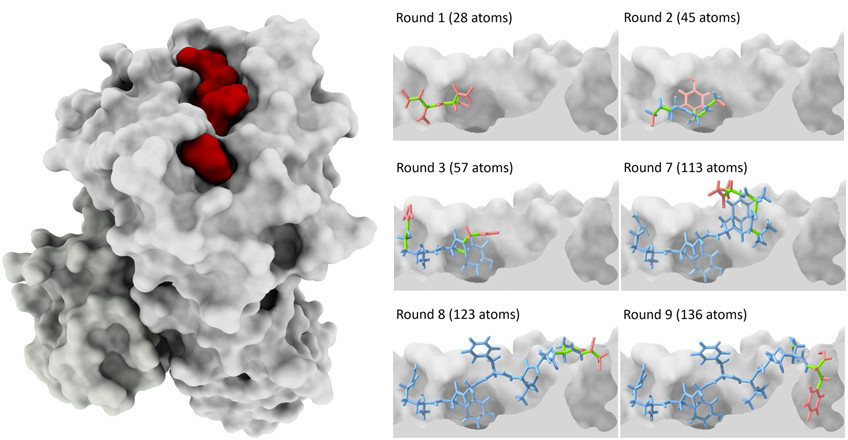 For predicting binding modes, we have worked on molecular docking. Briefly, the goal of docking is to predict how a small ligand (like a drug) binds to a protein receptor. Docking is routinely used as part of drug discovery pipelines. The size and flexibility of the ligand, however, represent important challenges for this type of prediction. We have developed an incremental meta-docking approach (DINC) to allow structural prediction of large ligands, including peptides (see figure above). Peptides are very flexible ligands involved in many important sinalization pathways. Fast and accurate prediction of protein-peptide complexes has many important biomedical applications, including vaccine design and cancer immunotherapy.
For predicting binding modes, we have worked on molecular docking. Briefly, the goal of docking is to predict how a small ligand (like a drug) binds to a protein receptor. Docking is routinely used as part of drug discovery pipelines. The size and flexibility of the ligand, however, represent important challenges for this type of prediction. We have developed an incremental meta-docking approach (DINC) to allow structural prediction of large ligands, including peptides (see figure above). Peptides are very flexible ligands involved in many important sinalization pathways. Fast and accurate prediction of protein-peptide complexes has many important biomedical applications, including vaccine design and cancer immunotherapy.
- Metabolic networks
-
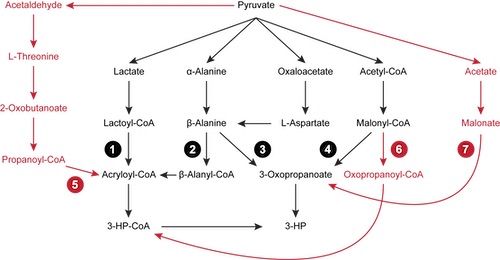 Metabolic engineering is the scientific process of manipulating the metabolism of a microorganism to produce valuable compounds. There have been numerous successes of metabolic engineering, including the well publicized biosynthesis of artemisinic acid, a precursor to the antimalarial drug artemisinin, and thebaine, a precursor to hydrocodone and morphine. We have developed pathfinding algorithms that facilitate the discovery of novel pathways using databases of known reactions and compounds.
Metabolic engineering is the scientific process of manipulating the metabolism of a microorganism to produce valuable compounds. There have been numerous successes of metabolic engineering, including the well publicized biosynthesis of artemisinic acid, a precursor to the antimalarial drug artemisinin, and thebaine, a precursor to hydrocodone and morphine. We have developed pathfinding algorithms that facilitate the discovery of novel pathways using databases of known reactions and compounds.
- Functional annotation
-
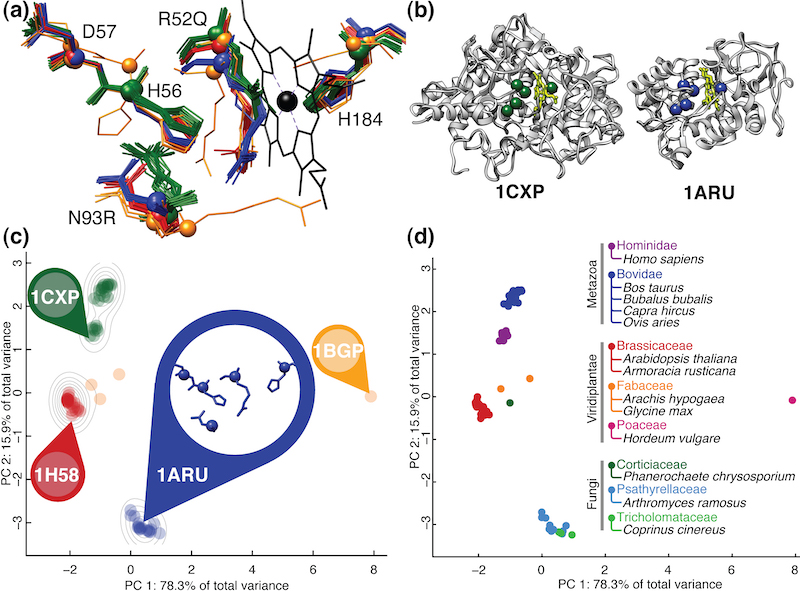 The function of a protein can often be characterized by a structural motif: a spatial arrangement of a certain number of residues that is unique to that function. We have developed efficient algorithms to search all known protein structures for occurences of a putative motif. We have also shown that families of proteins that share the same function often exhibit subtle patterns of structural variation that, using a careful analysis, can be related to, e.g., phylogenetic distance (as in the figure shown above).
The function of a protein can often be characterized by a structural motif: a spatial arrangement of a certain number of residues that is unique to that function. We have developed efficient algorithms to search all known protein structures for occurences of a putative motif. We have also shown that families of proteins that share the same function often exhibit subtle patterns of structural variation that, using a careful analysis, can be related to, e.g., phylogenetic distance (as in the figure shown above).
- Fundamentals of protein modeling
-
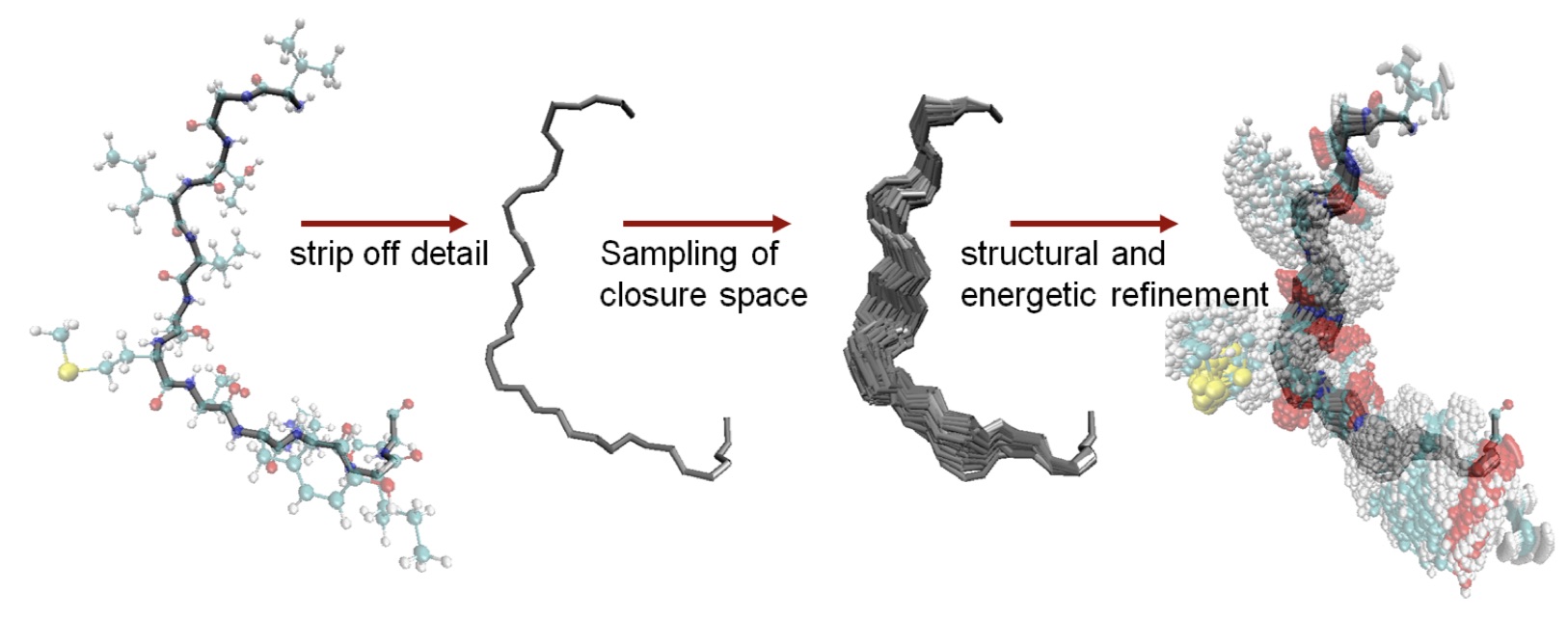 Proteins are not static structures. Accurately simulating the conformational changes in proteins is computationally very expensive. We have developed a variety of methods for generating (ensembles of) biophysically plausible conformations using techniques from robotics and geometric modelling.
Proteins are not static structures. Accurately simulating the conformational changes in proteins is computationally very expensive. We have developed a variety of methods for generating (ensembles of) biophysically plausible conformations using techniques from robotics and geometric modelling.
- Other topics
-
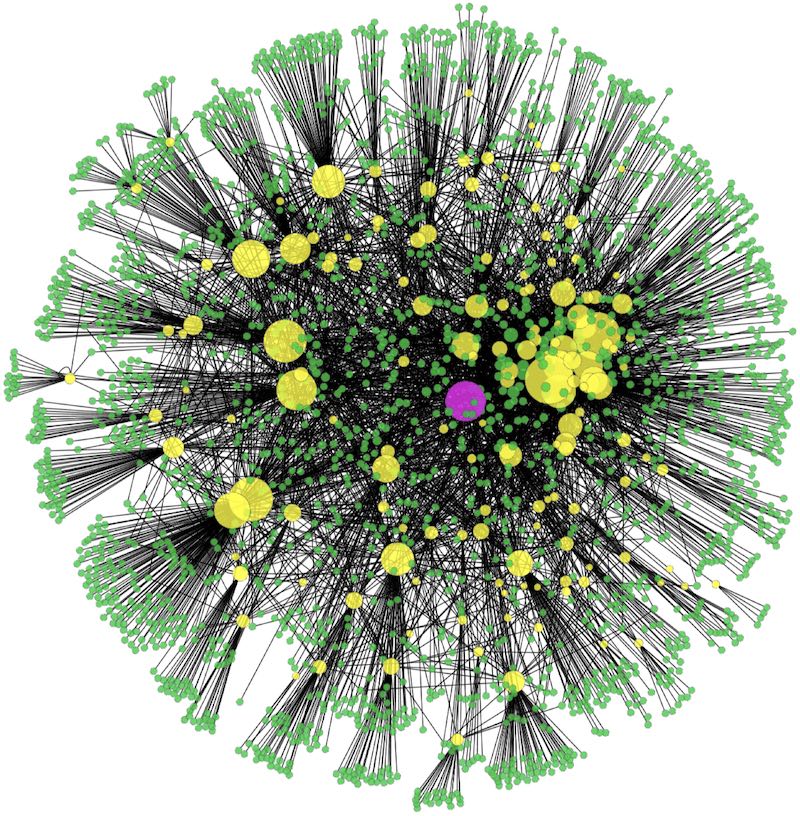 Similar to our computational robotics research, we are interested in a broad range of topics in computational biomedicine. We have worked with a broad range of people from the Texas Medical Center and elsewhere. The picture above shows a network of protein-protein interactions, taken from a review written by our group on the computational challenges in systems biology.
Similar to our computational robotics research, we are interested in a broad range of topics in computational biomedicine. We have worked with a broad range of people from the Texas Medical Center and elsewhere. The picture above shows a network of protein-protein interactions, taken from a review written by our group on the computational challenges in systems biology.